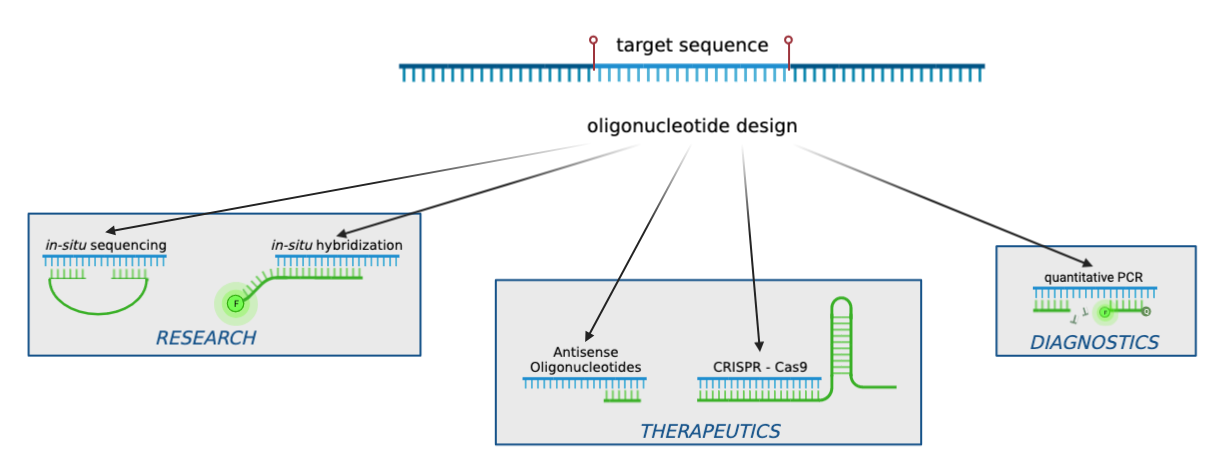
Oligonucleotides (abbrev. oligos) are short, synthetic strands of DNA or RNA that have many application areas, ranging from research to disease diagnosis or therapeutics. Oligos can be used as primers during DNA amplification, as probes for in situ hybridization or as guide RNAs for CRISPR-based gene editing. Based on the intended application and experimental design, researchers can customize the length, sequence composition, and thermodynamic properties of the designed oligos.
Oligo Designer Toolsuite provides ready-to-use oligo design pipelines for specific experimental setups, e.g. Padlock Probes for Spatial Transcriptomics.
News
- [2023.05.03] 🔥🔥 We will soon release a new version of the Oligo Designer Toolsuite with major updates! The new version 1) provides custom pipelines for MERFISH and SeqFISH+ probe design , 2) allows the user to design probes for all species available at NCBi or Ensemble and 3) allows the user to easily implement customized oligo design piplines using our new basic functionality modules! If you want to try out those functionalities already today, then check out the dev branch!
Installation
Requirements:
This package was build with Python 3.8 on ubuntu. It depends on the following additional tools Blast, BedTools, Bowtie and Bowtie2 that need to be installed independently. To install those tools via conda, please activate the Bioconda and conda-forge channels in your conda environment with and update conda and all packages in your environment:
conda config --add channels bioconda
conda config --add channels conda-forge
conda update conda
conda update --allFollow this instruction to install the required additional tools:
-
Blast (2.12 or higher) can be instelled via NCBI webpage or via Bioconda installation of Blast with:
conda install "blast>=2.12" -
BedTools (2.30 or higher) can be installed via BedTools GitHub or via Bioconda installation of BedTools with:
conda install "bedtools>=2.30" -
Bowtie (1.3 or higher) can be installed via Bowtie webpage or via Bioconda installation of Bowtie with:
conda install "bowtie>=1.3.1" -
Bowtie2 (2.5 or higher) can be installed via Bowtie2 webpage or via Bioconda installation of Bowtie2 with:
conda install "bowtie2>=2.5"
All other required packages are automatically installed if installation is done via :code:pip.
Install Options:
The installation of the package is done via pip. Note: if you are using conda, first install pip with: conda install pip.
PyPI install:
pip install oligo-designer-toolsuiteInstallation from source:
git clone https://github.com/HelmholtzAI-Consultants-Munich/oligo-designer-toolsuite.git-
Installation as python package (run inside directory):
pip install . -
Development Installation as python package (run inside directory):
pip install -e .[dev]
Implemented Oligo Design Pipelines
Padlock Probe Design
A padlock probe contains a constant backbone sequence of 53 nucleotides (nt) and the 5’- and 3’- arms, which are complementary to the corresponding mRNA sequence. The gene-specific arms of padlock probes are around 20nt long each, thus the total length of the gene-specific sequence of each padlock is 40nt.
Usage
Command-Line Call:
To create padlock probes you can run the pipeline with
padlock_probe_designer -c ./config/padlock_probe_designer.yaml -o output/ [-d False]where:
-c: config file, which contains parameter settings, specific to padlock probe design, ./config/padlock_probe_designer.yaml contains default parameter settings-o: output folder, where results of pipeline are storedannotationsfolder: downloaded gene and genome annotation as well as constructed transcriptomeprobesfolder: list of probes per gene, which fulfill user-defined criteria, given in config fileprobesetsfolder: sets of non-overlapping probes per gene, ranked by best set criteriapadlock_probesfolder: final padlock probe sequences per gene, ready to order
-d: optional, 'download only' option, where only gene and genome annotation files are downloaded but no probes generated, default: False
All steps and config parameters will be documented in a log file, that is saved in the directory where the pipeline is executed from. The logging file will have the format: log_padlock_probe_designer_{year}-{month}-{day}-{hour}-{minute}.txt.
Python Import:
Import padlock probe design pipeline as python package:
import oligo_designer_toolsuite.pipelines.padlock_probe_designer as packlock_probe_designer
config = './config/padlock_probe_designer.yaml'
dir_output = './padlock_probes'
annotations = packlock_probe_designer.download_annotations(config, dir_output)
packlock_probe_designer.filter_probes(config, annotations, dir_output)
del annotations # free memory
packlock_probe_designer.generate_probe_sets(config, dir_output)
packlock_probe_designer.design_padlock_probes(config, dir_output)Contributing
Contributions are more than welcome! Everything from code to notebooks to examples and documentation are all equally valuable so please don't feel you can't contribute. To contribute please fork the project make your changes and submit a pull request. We will do our best to work through any issues with you and get your code merged into the main branch.
How to cite
If the Ologo Designer Toolsuite is useful for your research, consider citing the package:
@software{campi_2023_7823048,
author = { Isra Mekki,
Francesco Campi,
Louis Kümmerle,
Chelsea Bright,
Malte Lücken
Fabian Theis,
Marie Piraud,
Lisa Barros de Andrade e Sousa
},
title = {{Oligo Designer Toolsuite}},
year = 2023,
publisher = {Zenodo},
version = {v0.1.3},
doi = {10.5281/zenodo.7823048},
url = {https://doi.org/10.5281/zenodo.7823048}
}If you are using one of the spatial transcriptomics pipelines provided along the Oligo Designer Toolsuite, consider citing the paper:
@article {Kuemmerle2022.08.16.504115,
author = { Louis B. Kuemmerle,
Malte D. Luecken,
Alexandra B. Firsova
Lisa Barros de Andrade e Sousa
Lena Strasser
Lukas Heumos
Ilhem Isra Mekki
Krishnaa T. Mahbubani
Alexandros Sountoulidis
Tamas Balassa
Ferenc Kovacs
Peter Horvath
Marie Piraud
Ali Ertürk
Christos Samakovlis
Fabian J. Theis},
title = {{Probe set selection for targeted spatial transcriptomics}},
year = {2022},
publisher = {Cold Spring Harbor Laboratory},
journal = {bioRxiv},
doi = {10.1101/2022.08.16.504115},
URL = {https://www.biorxiv.org/content/early/2022/08/17/2022.08.16.504115}
}License
oligo-designer-toolsuite is released under the MIT license. See LICENSE for additional details about it.