
GA4GH NCBI API
Genomic data is often very large and requires metadata in order to be used as part of an inquiry. To make the process of discovering and analyzing genomic data less of a challenge, the Global Alliance for Genomics and Health (GA4GH) has designed an "easy-to-implement" HTTP API that lets you get at just the data relevant to a specific inquiry.
The National Center for Biotechnology Information (NCBI) curates a great deal of invaluable genomic data. By making these data available using GA4GH methods the NCBI data becomes discoverable using GA4GH HTTP Clients.
This software was developed as part of the March 2017 NCBI Hackathon. A presentation and draft manuscript are available, which will be submitted to F1000.
Installation
Install Python 2
This application runs in Python 2.7. It can be installed using this command.
sudo apt-get install python-dev python-virtualenv zlib1g-dev libxslt1-dev
Get the code
The following commands will download the latest code available in this
repository, enters the directory and installs the package. It will make
available in your current Python environment the ncbi and ga4gh_ncbi
modules behind this application.
git clone https://github.com/NCBI-Hackathons/ga4gh-ncbi-api.git
cd ga4gh-ncbi-api
pip install .Installing NGS
This software makes use of the NCBI NGS Python bindings, which currently must be downloaded and installed on the host system.
During the first run of your application, depending on the configuration, the library may take a few minutes to download. If your application is not responsive at first, this may be the cause.
Their most recent downloads are available here.
Running via docker
Docker pull
sudo docker build . -t ga4gh-ncbi-api
sudo docker run ga4gh-ncbi-api -d -p 8000:80
Example usage
We will provide a iPython Notebook that demonstrates interacting with this software.
Architecture
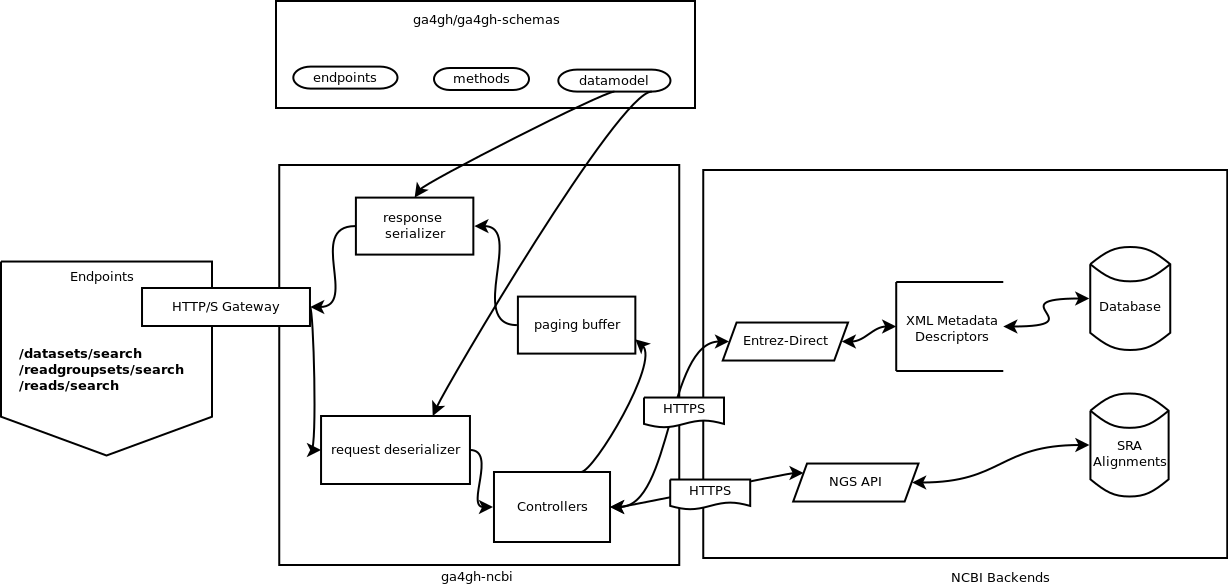
GA4GH Schemas
The GA4GH has designed a schema in Google Protocol Buffers which provides the data serialization and de-serialization layers for this application.
What is Protocol Buffers?
Protocol Buffers is an interchange format Open Sourced by Google. It allows schemas to be defined in a language neutral IDL. Bindings can be generated for your language of choice, making available prototypical messages that can be "filled out" by implementors.
This allows a portable template to be used by our software. This server also uses serialization helpers made available by the ga4gh-schemas python module. Once an NCBI message has been mapped to the protocol buffers, the resulting message can be reliably converted to and from JSON.
Dataset Service
Allows one to interrogate about the containing project used for collecting a genomics dataset.
Read Service
Allows one to interrogate about the metadata regarding a run, as well as the underlying alignments.
NCBI APIs
The NCBI provides multiple ways to interrogate about data they provide.
edirect
Returns XML metadata about projects, runs, and samples.
NGS
Allows one to interrogate a run about aligned reads.