A working conda environment could be downloaded. Please follow the README.txt file there to activate this environment. Once successfully activated, you may skip the installation steps.
News Board
2022/05/03 New features experimented, details introduced in
notebooks2022/04/29 VeloAE updated to 0.2.0. To simplify the project, the folder
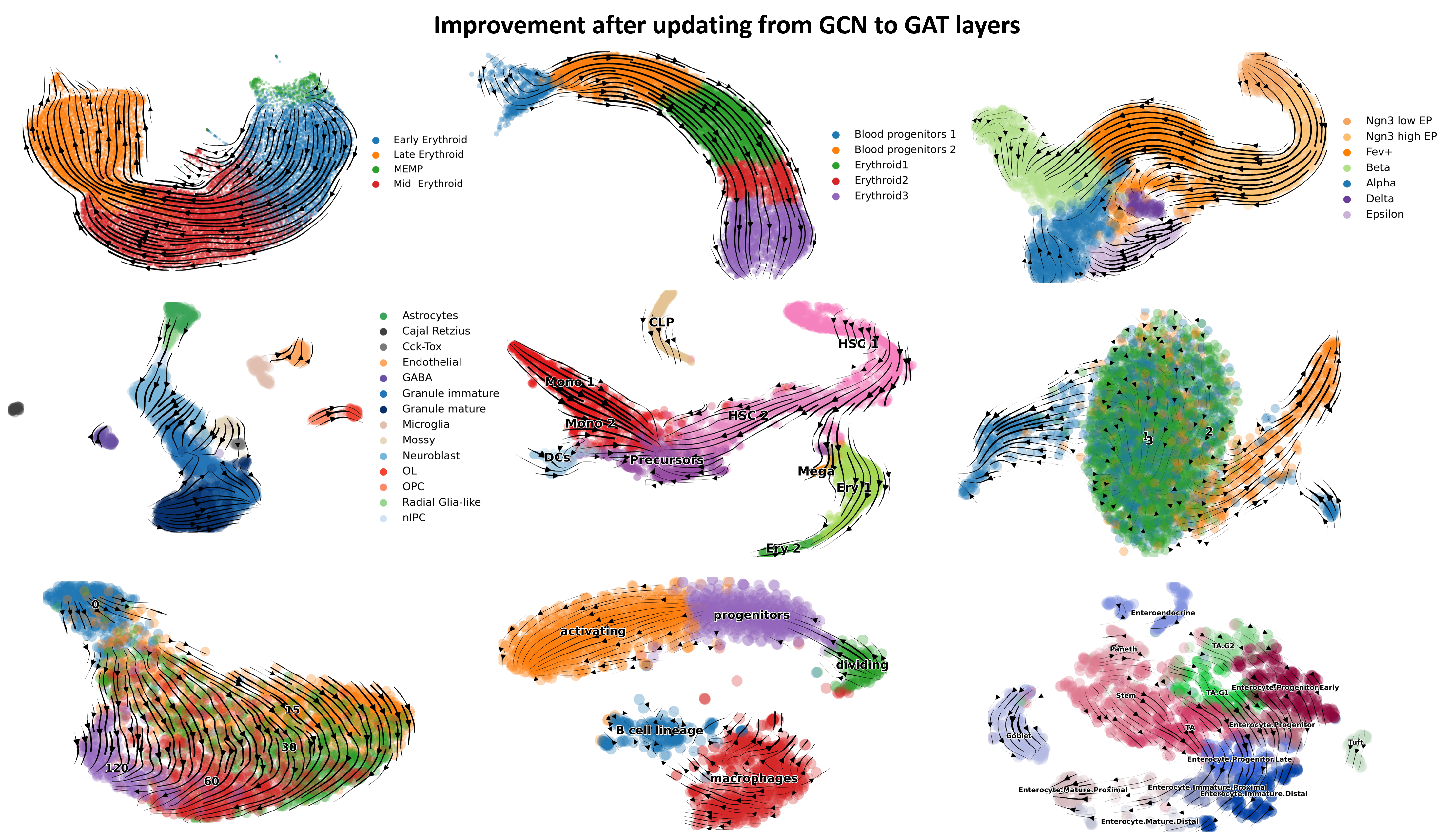
notebooksis reorganized and only notebooks involving veloAE experiments are kept, scvelo dynamical mode models are additionally included for comparison. The previous data are backuped in the branchpaper-version-backup2022/04/29 Version Updating in progress. We thank @Mingze Yuan from PKU for his great insights in correcting issues regarding veloAE's cohort aggregation module and a suggestion on replacing GCN with GAT layers, which leads to better performances on challenging datasets like human and mouse bonemarrow, a preview of updated results:
2022/04/04 Exciting news! UnitTVelo, a new single cell RNA velocity estimation tool that addresses the challenging datasets of existing tools is published by our lab.
VeloAE
Low-dimensional Estimation of Single Cell RNA Velocity with AutoEncoder.
VeloAE can learn low-dimensional projections for count matrices leveraging a tailored AutoEncoder, with the aim to obtain better representations for RNA velocity estimation. Results of VeloAE could be previewed in the jupyter notebooks located under the notebooks folder.
Our revised manuscript is in progress, while the first version could be found in bioarxiv with the title Representation learning of RNA velocity reveals robust cell transitions
Follow https://github.com/qiaochen/VeloAE/blob/main/notebooks/readme.md to access the datasets used in the notebooks and manuscript
Install
Dependencies:
- numpy
- scipy
- pandas
- matplotlib
- scikit-learn
- anndata>=0.7.4
- scanpy>=1.5.1
- scvelo>=0.2.2
- pytorch>=1.7
- pytorch-geometric>=1.6.3
Install using pip
pip install git+https://github.com/qiaochen/VeloAEAn example local installation procedure in linux system
conda create -n veloAE
conda activate veloAE
git clone https://github.com/qiaochen/VeloAE.git
cd VeloAE
conda install python=3.7
pip3 install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cu113
pip install torch-scatter -f https://data.pyg.org/whl/torch-1.11.0+cu113.html
pip install torch-sparse -f https://data.pyg.org/whl/torch-1.11.0+cu113.html
pip install torch-geometric
pip install .Usage
Command line usage
Basic execution command and arguments:
Project velocity into low-dimensional space using a trained model.
veloproj --refit false --vis_type_col clusters --scv_n_jobs 10 --vis-key X_umap --nb_g_src X --gumbsoft_tau 5 --fit_offset_pred true --adata notebooks/dentategyrus/data/DentateGyrus/10X43_1.h5ad --device cuda:3 --model-name ./notebooks/dentategyrus/dentategyrus_model.cpt-
Arguments:
- --refit: if false, do not fit a new model, should specify an existing model for projection.
- --vis_type_col: The column name in adata.obs Dataframe for the cell type/cluster column, for result visualization.
- --scv_n_jobs: Number of cores used for scvelo operations.
- --vis-key: The key in adata.obsm for embeddings, e.g. 'X_umap'.
- --nb_g_src: expression matrix for generating neighborhood graph of cells, can be 'X' (transcriptome), 'S' (spliced), and 'U' (unspliced).
- --gumbsoft_tau: temperature parameter of gumbel softmax function, a smaller value (e.g., 1) makes attention sparse and training more challenging, while a larger value (e.g., 10) makes attention more evenly distributed and loss more smoothly converge to lower value.
- --fit_offset_pred: whether or not to fit offset when estimating velocity using linear regression in projected low-dimensional space
- --adata: path to the Anndata with X_umap, transcriptom, spliced and unspliced mRNA expressions.
- --device: gpu or cpu (e.g., cuda:2). Fitting using GPU is much faster than cpu.
- --model-name: path to the existing model.
-
Output:
- A new low-dimensional Anndata object with default name "projection.h5ad" in the output folder (default ./)
- An uncolored figure showing the low-dimensional velocity field, stored in folder './figures'.
Fit a new veloAE model and project velocity into low-dimensional space.
veloproj --lr 1e-5 --nb_g_src X --gumbsoft_tau 5 --fit_offset_pred true --vis_type_col clusters --scv_n_jobs 10 --vis-key X_umap --refit true --adata notebooks/dentategyrus/data/DentateGyrus/10X43_1.h5ad --device cuda:3 --model-name dentategyrus_model.cpt --output './' -
Arguments:
- --lr: learning rate, (tunning it if the model does not learn given the default configuration).
- --nb_g_src: expression matrix for generating neighborhood graph of cells, can be 'X' (transcriptome), 'S' (spliced), and 'U' (unspliced).
- --gumbsoft_tau: temperature parameter of gumbel softmax function, a smaller value (e.g., 1) makes attention sparse and training more challenging, while a larger value (e.g., 10) makes attention more evenly distributed and loss more smoothly converge to lower value.
- --fit_offset_pred: whether or not to fit offset when estimating velocity using linear regression in projected low-dimensional space
- --vis_type_col: The column name in adata.obs Dataframe for the cell type/cluster column, for result visualization.
- --scv_n_jobs: Number of cores used for scvelo operations.
- --vis-key: The key in adata.obsm for embeddings, e.g. 'X_umap'.
- --refit: if true, fit a new model.
- --adata: path to the Anndata with X_umap, transcriptom, spliced and unspliced mRNA expressions.
- --device: gpu or cpu. Fitting using GPU is much faster than cpu.
- --model-name: path for storing the trained model.
- --output: specify the directory for storing fitting results.
-
Output:
- A trained model with "--model-name" in the output folder (default ./, can be specified using arg --output).
- A new low-dimensional Anndata instance with default name "projection.h5ad" in the output folder (default ./, can be specified using arg --output)
- An uncolored/colored figure showing the low-dimensional velocity field, stored in folder './figures'.
- A plot showing the training loss curve named as
training_loss.pngin the output folder (default ./, can be specified using arg --output).

If the training loss decreases smoothly but not converge after exhausting all the epochs, please try either more epochs or larger learning rates. Note, however, that learning rates should not be set too large to make the loss osciliates and never decreases.
Use command line help to investigate more arguments.
veloproj -h