[TOC]
LocARNA: Alignment of RNAs
The LocARNA package provides several tools for the structural analysis of RNA. LocARNA's main functionality is to align a set of a priori unaligned RNAs sequences and at the same time predict their common structure. In this way, LocARNA performs simultaneous alignment and folding in the spirit of the classical Sankoff algorithm, but implements strategies to perform this computationally challenging task efficiently and comparably fast.
Due to the central ability to simultaneously assess sequence similarity and the similarity of predicted structure, LocARNA is recommends itself for the analysis of RNAs in the twilight zone (around or below 60% sequence identity), where alignments based on only sequence similarity are unreliable. Thus, it could be sometimes easier and typically faster to align highly similar RNAs using pure sequence alignment tools; similarily, RNAs with existing trusted alignments can be more efficiently analyzed based on specialized tools like RNAalifold, R-scape, or Infernal.
Example of standard usage
Most of the package's functionality is accessible via the command-line tool mlocarna
through its various options. In the simplest case, we provide the input
sequences in a fasta file.
$ mlocarna archaea.fayields text output and writes results (and intermediary results) to disk; here to folder archaea.out.
As main result, it produces the alignment of the seven short RNA sequences in archaea.fa together with a consensus structure:
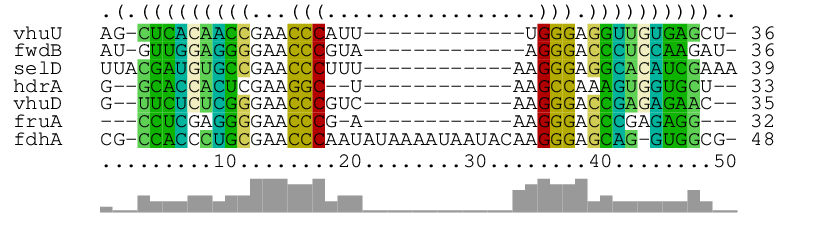
The graphical RNAalifold-generated output shows the aligned RNAs (with gaps), the consensus structure as dot-bracket string on top, and the column similarities by bars at the bottom. In the way of alifold, columns are color-coded to visualize compensatory and incompatible mutations at predicted base pairs.
More on features and alignment variants
LocARNA distinguishes itself from many other Sankoff-style multiple alignment programs by its high performance (strongly improved in the 2.x line) and low memory complexity, high accuracy, and a broad set of features. As unique features, it offers structure-local alignment, flexible structure constraints and anchor constraints, specialized realignment modes for refining existing alignments, and provides efficient computation of reliabilities in sequence-structure alignment. The package offers a robust core of features and is used as experimental platform for new RNA alignment related methods.
Multiple alignment can be performed in one of several different ways:
-
progressive alignment using sequence-structure alignment of profiles
-
progressive alignment after consistency transformation using T-Coffee
-
progressive alignment using probabilistic consistency transformation and sequence-structure profile alignments, optionally followed by iterative refinement.
Besides of global alignment, LocARNA supports two kinds of locality. Local alignment as it is known from sequence alignment, identifies and aligns the best matching subsequences. This form of locality is called sequence local to distinguish it from structural locality. When performing structure local alignment, LocARNA identifies and aligns the best matching substructures in the RNAs. The sequences of those substructures can be discontinuous on the sequence level, but remain connected via structural bonds.
Alignment Reliabilities (LocARNA-P). In this special, probabilistic mode of operation LocARNA supports the efficient computation of match probabilities, probabilistic consistency transformation for more accurate multiple alignment, and generates reliability profiles of multiple alignments.
Installation
The software can be installed on recent Linux or MacOSX systems; Windows is untested but should be supported via WSL.
Installation from Conda package (recommended)
On Mac/Linux, LocARNA is installed most easily via Conda from a pre-compiled package. For this purpose, install Conda and run from the command line:
conda install -c conda-forge -c bioconda locarnaAlternative installation from source
Installing from source requires a C++ compiler (GNU C++, Clang, ...) and Autotools. Moreover, it depends on the Vienna RNA package.
Installation from source distribution
Obtain the tar.gz source distribution, e.g. from Github
https://github.com/s-will/LocARNA/releases
Then, build and install like
tar xzf locarna-xxx.tar.gz
cd locarna-xxx
./configure --prefix=/usr/local
make
make installIs Vienna RNA installed in a non-standard location, this has to be
specified by configure option `--with-vrna=path-to-vrna.
Installing from source furthermore allows testing via
make checkand building documentation locally by
make doxygen-docBuilding documentation requires additional tools: doxygen, pod2markdown and pandoc.
Installation from the git repository
Installing from repository is possible after cloning and setting up the autotools suite. This is most easily achieved by running
autoreconf -iin the cloned repository. Then, the installation essentially works like installing from source distribution. Note that, we will however require additional tools to build the documentation: help2man, pod2man.
Usage
For instructions on the use of the tools, please see the documentation / man pages of the single tools
-
mlocarna --- for multiple alignment of RNAs. This program supports most of the functionality of the package via a high level interface.
-
locarna --- for pairwise alignment
-
locarna_p --- for pairwise computation of alignment partition function and (sequence and structure) match probabilities
-
sparse --- for structurally stronger sparsified pairwise alignment
For additional functionality and special purposes, see
-
exparna_p --- for generating exact matches from the ensembles of two RNAs
-
locarnate --- for multiple alignment of RNAs via T-Coffee. This script offers multiple alignment of RNAs that is performed by sequence-structurally aligning all pairs of RNAs and then using T-Coffee to construct a common multiple alignment out of all pairwise ones.
Web server
The core functionality of the package is accessible through a web interface at
http://rna.informatik.uni-freiburg.de
Contact
Main author and contact: Sebastian Will sebastian.will (at) polytechnique.edu
References
- Sebastian Will, Kristin Reiche, Ivo L. Hofacker, Peter F. Stadler, and Rolf Backofen. Inferring non-coding RNA families and classes by means of genome-scale structure-based clustering. PLOS Computational Biology, 3 no. 4 pp. e65, 2007. doi:10.1371/journal.pcbi.0030065
- Sebastian Will, Tejal Joshi, Ivo L. Hofacker, Peter F. Stadler, and Rolf Backofen. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA, 18 no. 5 pp. 900-914, 2012. doi:10.1261/rna.029041.111
- Sebastian Will, Michael Yu, and Bonnie Berger. Structure-based Whole Genome Realignment Reveals Many Novel Non-coding RNAs. Genome Research, no. 23 pp. 1018-1027, 2013. doi:10.1101/gr.137091.111
- Sebastian Will, Christina Otto, Milad Miladi, Mathias Mohl, and Rolf Backofen. SPARSE: quadratic time simultaneous alignment and folding of RNAs without sequence-based heuristics. Bioinformatics, 31(15):2489–2496, 2015. doi:10.1093/bioinformatics/btv185