Cell Detection
⭐ Showcase
NeurIPS 22 Cell Segmentation Competition
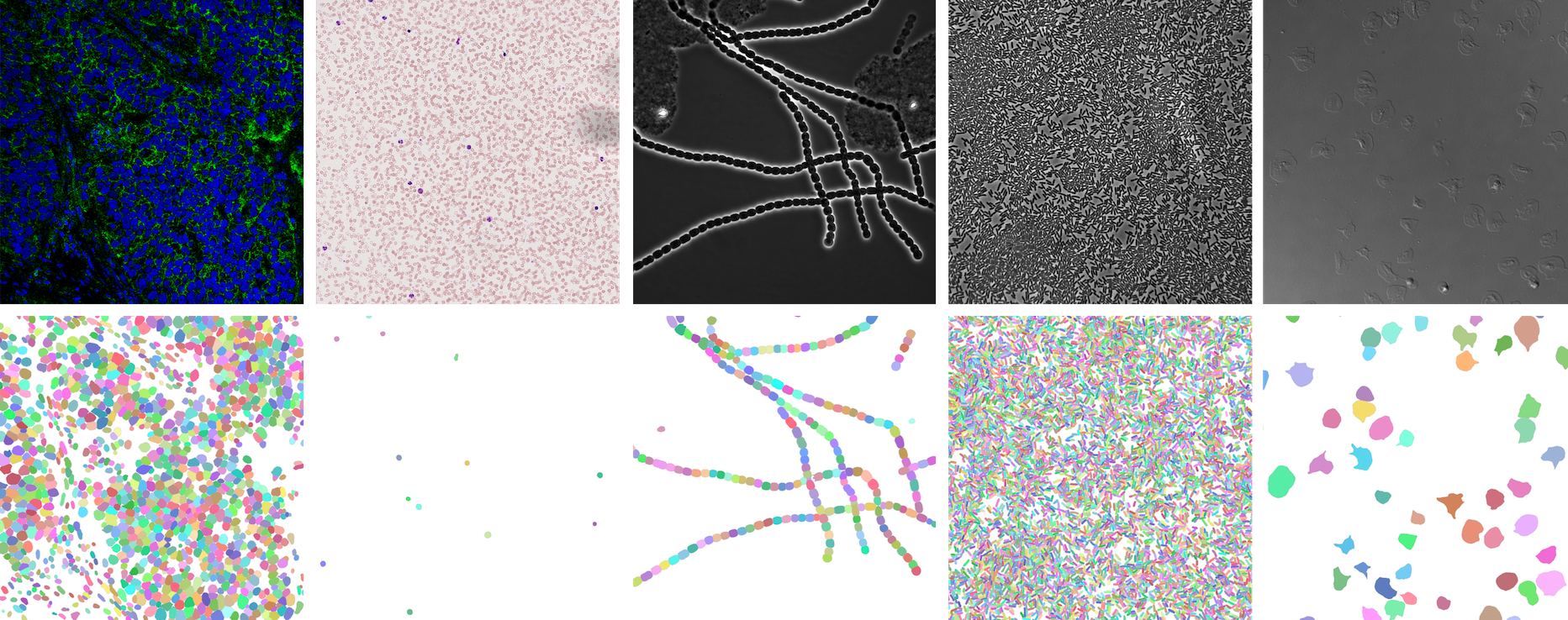 https://openreview.net/forum?id=YtgRjBw-7GJ
https://openreview.net/forum?id=YtgRjBw-7GJ
Nuclei of U2OS cells in a chemical screen
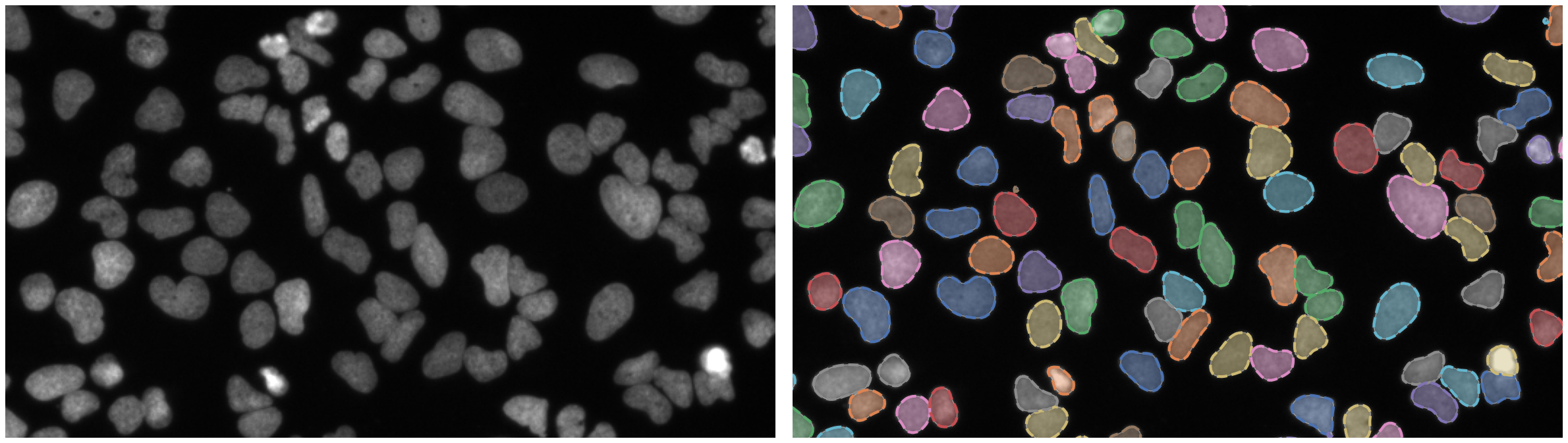 https://bbbc.broadinstitute.org/BBBC039 (CC0)
https://bbbc.broadinstitute.org/BBBC039 (CC0)
P. vivax (malaria) infected human blood
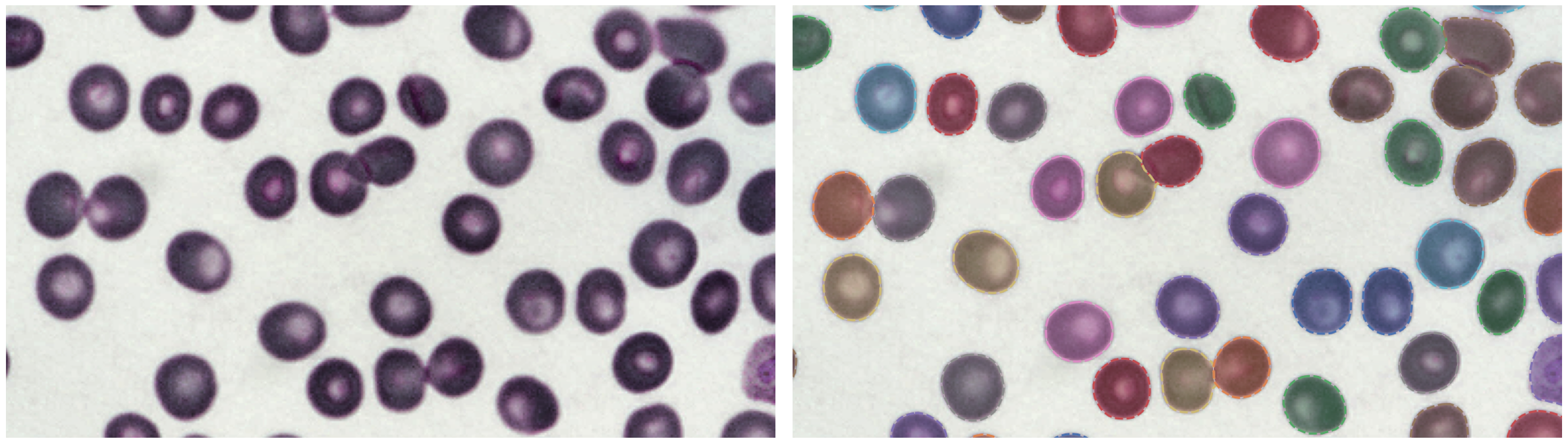 https://bbbc.broadinstitute.org/BBBC041 (CC BY-NC-SA 3.0)
https://bbbc.broadinstitute.org/BBBC041 (CC BY-NC-SA 3.0)
🛠 Install
Make sure you have PyTorch installed.
PyPI
pip install -U celldetectionGitHub
pip install git+https://github.com/FZJ-INM1-BDA/celldetection.git💾 Trained models
model = cd.fetch_model(model_name, check_hash=True)| model name | training data | link |
|---|---|---|
ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c |
BBBC039, BBBC038, Omnipose, Cellpose, Sartorius - Cell Instance Segmentation, Livecell, NeurIPS 22 CellSeg Challenge | 🔗 |
Run a demo with a pretrained model
```python import torch, cv2, celldetection as cd from skimage.data import coins from matplotlib import pyplot as plt # Load pretrained model device = 'cuda' if torch.cuda.is_available() else 'cpu' model = cd.fetch_model('ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c', check_hash=True).to(device) model.eval() # Load input img = coins() img = cv2.cvtColor(img, cv2.COLOR_GRAY2RGB) print(img.dtype, img.shape, (img.min(), img.max())) # Run model with torch.no_grad(): x = cd.to_tensor(img, transpose=True, device=device, dtype=torch.float32) x = x / 255 # ensure 0..1 range x = x[None] # add batch dimension: Tensor[3, h, w] -> Tensor[1, 3, h, w] y = model(x) # Show results for each batch item contours = y['contours'] for n in range(len(x)): cd.imshow_row(x[n], x[n], figsize=(16, 9), titles=('input', 'contours')) cd.plot_contours(contours[n]) plt.show() ```🔬 Architectures
import celldetection as cdContour Proposal Networks
- [`cd.models.CPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CPN) - [`cd.models.CpnU22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnU22) - [`cd.models.CPNCore`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CPNCore) - [`cd.models.CpnResUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResUNet) - [`cd.models.CpnSlimU22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnSlimU22) - [`cd.models.CpnWideU22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnWideU22) - [`cd.models.CpnResNet18FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet18FPN) - [`cd.models.CpnResNet34FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet34FPN) - [`cd.models.CpnResNet50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet50FPN) - [`cd.models.CpnResNeXt50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt50FPN) - [`cd.models.CpnResNet101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet101FPN) - [`cd.models.CpnResNet152FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet152FPN) - [`cd.models.CpnResNet18UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet18UNet) - [`cd.models.CpnResNet34UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet34UNet) - [`cd.models.CpnResNet50UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet50UNet) - [`cd.models.CpnResNeXt101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt101FPN) - [`cd.models.CpnResNeXt152FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt152FPN) - [`cd.models.CpnResNeXt50UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt50UNet) - [`cd.models.CpnResNet101UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet101UNet) - [`cd.models.CpnResNet152UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNet152UNet) - [`cd.models.CpnResNeXt101UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt101UNet) - [`cd.models.CpnResNeXt152UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnResNeXt152UNet) - [`cd.models.CpnWideResNet50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnWideResNet50FPN) - [`cd.models.CpnWideResNet101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnWideResNet101FPN) - [`cd.models.CpnMobileNetV3LargeFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnMobileNetV3LargeFPN) - [`cd.models.CpnMobileNetV3SmallFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnMobileNetV3SmallFPN)PyTorch Image Models (timm)
Also have a look at [Timm Documentation](https://huggingface.co/docs/timm/index). ```python import timm timm.list_models(filter='*') # explore available models ``` - [`cd.models.CpnTimmMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnTimmMaNet) - [`cd.models.CpnTimmUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnTimmUNet) - [`cd.models.TimmEncoder`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.timmodels.TimmEncoder) - [`cd.models.TimmFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.TimmFPN) - [`cd.models.TimmMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.manet.TimmMaNet) - [`cd.models.TimmUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.TimmUNet)Segmentation Models PyTorch (smp)
```python import segmentation_models_pytorch as smp smp.encoders.get_encoder_names() # explore available models ``` ```python encoder = cd.models.SmpEncoder(encoder_name='mit_b5', pretrained='imagenet') ``` Find a list of [Smp Encoders](https://smp.readthedocs.io/en/latest/encoders.html) in the `smp` documentation. - [`cd.models.CpnSmpMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnSmpMaNet) - [`cd.models.CpnSmpUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.cpn.CpnSmpUNet) - [`cd.models.SmpEncoder`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.smp.SmpEncoder) - [`cd.models.SmpFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.SmpFPN) - [`cd.models.SmpMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.manet.SmpMaNet) - [`cd.models.SmpUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.SmpUNet)U-Nets
```python # U-Nets are available in 2D and 3D import celldetection as cd model = cd.models.ResNeXt50UNet(in_channels=3, out_channels=1, nd=3) ``` - [`cd.models.U22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.U22) - [`cd.models.U17`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.U17) - [`cd.models.U12`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.U12) - [`cd.models.UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.UNet) - [`cd.models.WideU22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.WideU22) - [`cd.models.SlimU22`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.SlimU22) - [`cd.models.ResUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResUNet) - [`cd.models.UNetEncoder`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.UNetEncoder) - [`cd.models.ResNet50UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNet50UNet) - [`cd.models.ResNet18UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNet18UNet) - [`cd.models.ResNet34UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNet34UNet) - [`cd.models.ResNet152UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNet152UNet) - [`cd.models.ResNet101UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNet101UNet) - [`cd.models.ResNeXt50UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNeXt50UNet) - [`cd.models.ResNeXt152UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNeXt152UNet) - [`cd.models.ResNeXt101UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.ResNeXt101UNet) - [`cd.models.WideResNet50UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.WideResNet50UNet) - [`cd.models.WideResNet101UNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.WideResNet101UNet) - [`cd.models.MobileNetV3SmallUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.MobileNetV3SmallUNet) - [`cd.models.MobileNetV3LargeUNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.unet.MobileNetV3LargeUNet)MA-Nets
```python # Many MA-Nets are available in 2D and 3D import celldetection as cd encoder = cd.models.ConvNeXtSmall(in_channels=3, nd=3) model = cd.models.MaNet(encoder, out_channels=1, nd=3) ``` - [`cd.models.MaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.manet.MaNet) - [`cd.models.SmpMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.manet.SmpMaNet) - [`cd.models.TimmMaNet`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.manet.TimmMaNet)Feature Pyramid Networks
- [`cd.models.FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.FPN) - [`cd.models.ResNet18FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNet18FPN) - [`cd.models.ResNet34FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNet34FPN) - [`cd.models.ResNet50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNet50FPN) - [`cd.models.ResNeXt50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNeXt50FPN) - [`cd.models.ResNet101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNet101FPN) - [`cd.models.ResNet152FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNet152FPN) - [`cd.models.ResNeXt101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNeXt101FPN) - [`cd.models.ResNeXt152FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.ResNeXt152FPN) - [`cd.models.WideResNet50FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.WideResNet50FPN) - [`cd.models.WideResNet101FPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.WideResNet101FPN) - [`cd.models.MobileNetV3LargeFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.MobileNetV3LargeFPN) - [`cd.models.MobileNetV3SmallFPN`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.fpn.MobileNetV3SmallFPN)ConvNeXt Networks
```python # ConvNeXt Networks are available in 2D and 3D import celldetection as cd model = cd.models.ConvNeXtSmall(in_channels=3, nd=3) ``` - [`cd.models.ConvNeXt`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.convnext.MaNet) - [`cd.models.ConvNeXtTiny`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.convnext.ConvNeXtTiny) - [`cd.models.ConvNeXtSmall`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.convnext.ConvNeXtSmall) - [`cd.models.ConvNeXtBase`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.convnext.ConvNeXtBase) - [`cd.models.ConvNeXtLarge`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.convnext.ConvNeXtLarge)Residual Networks
```python # Residual Networks are available in 2D and 3D import celldetection as cd model = cd.models.ResNet50(in_channels=3, nd=3) ``` - [`cd.models.ResNet18`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNet18) - [`cd.models.ResNet34`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNet34) - [`cd.models.ResNet50`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNet50) - [`cd.models.ResNet101`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNet101) - [`cd.models.ResNet152`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNet152) - [`cd.models.WideResNet50_2`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.WideResNet50_2) - [`cd.models.ResNeXt50_32x4d`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNeXt50_32x4d) - [`cd.models.WideResNet101_2`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.WideResNet101_2) - [`cd.models.ResNeXt101_32x8d`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNeXt101_32x8d) - [`cd.models.ResNeXt152_32x8d`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.resnet.ResNeXt152_32x8d)Mobile Networks
- [`cd.models.MobileNetV3Large`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.mobilenetv3.MobileNetV3Large) - [`cd.models.MobileNetV3Small`](https://docs.celldetection.org/en/latest/celldetection.models.html#celldetection.models.mobilenetv3.MobileNetV3Small)🐳 Docker
Find us on Docker Hub: https://hub.docker.com/r/ericup/celldetection
You can pull the latest version of celldetection via:
docker pull ericup/celldetection:latestCPN inference via Docker with GPU
``` docker run --rm \ -v $PWD/docker/outputs:/outputs/ \ -v $PWD/docker/inputs/:/inputs/ \ -v $PWD/docker/models/:/models/ \ --gpus="device=0" \ celldetection:latest /bin/bash -c \ "python cpn_inference.py --tile_size=1024 --stride=768 --precision=32-true" ```CPN inference via Docker with CPU
``` docker run --rm \ -v $PWD/docker/outputs:/outputs/ \ -v $PWD/docker/inputs/:/inputs/ \ -v $PWD/docker/models/:/models/ \ celldetection:latest /bin/bash -c \ "python cpn_inference.py --tile_size=1024 --stride=768 --precision=32-true --accelerator=cpu" ```Apptainer
You can also pull our Docker images for the use with Apptainer (formerly Singularity) with this command:
apptainer pull --dir . --disable-cache docker://ericup/celldetection:latest🤗 Hugging Face Spaces
Find us on Hugging Face and upload your own images for segmentation: https://huggingface.co/spaces/ericup/celldetection
There's also an API (Python & JavaScript), allowing you to utilize community GPUs (currently Nvidia A100) remotely!
Hugging Face API
### Python ```python from gradio_client import Client # Define inputs (local filename or URL) inputs = 'https://raw.githubusercontent.com/scikit-image/scikit-image/main/skimage/data/coins.png' # Set up client client = Client("ericup/celldetection") # Predict overlay_filename, img_filename, h5_filename, csv_filename = client.predict( inputs, # str: Local filepath or URL of your input image # Model name 'ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c', # Custom Score Threshold (numeric value between 0 and 1) False, .9, # bool: Whether to use custom setting; float: Custom setting # Custom NMS Threshold False, .3142, # bool: Whether to use custom setting; float: Custom setting # Custom Number of Sample Points False, 128, # bool: Whether to use custom setting; int: Custom setting # Overlapping objects True, # bool: Whether to allow overlapping objects # API name (keep as is) api_name="/predict" ) # Example usage: Code below only shows how to use the results from matplotlib import pyplot as plt import celldetection as cd import pandas as pd # Read results from local temporary files img = imread(img_filename) overlay = imread(overlay_filename) # random colors per instance; transparent overlap properties = pd.read_csv(csv_filename) contours, scores, label_image = cd.from_h5(h5_filename, 'contours', 'scores', 'labels') # Optionally display overlay cd.imshow_row(img, img, figsize=(16, 9)) cd.imshow(overlay) plt.show() # Optionally display contours with text cd.imshow_row(img, img, figsize=(16, 9)) cd.plot_contours(contours, texts=['score: %d%%\narea: %d' % s for s in zip((scores * 100).round(), properties.area)]) plt.show() ``` ### Javascript ```javascript import { client } from "@gradio/client"; const response_0 = await fetch("https://raw.githubusercontent.com/scikit-image/scikit-image/main/skimage/data/coins.png"); const exampleImage = await response_0.blob(); const app = await client("ericup/celldetection"); const result = await app.predict("/predict", [ exampleImage, // blob: Your input image // Model name (hosted model or URL) "ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c", // Custom Score Threshold (numeric value between 0 and 1) false, .9, // bool: Whether to use custom setting; float: Custom setting // Custom NMS Threshold false, .3142, // bool: Whether to use custom setting; float: Custom setting // Custom Number of Sample Points false, 128, // bool: Whether to use custom setting; int: Custom setting // Overlapping objects true, // bool: Whether to allow overlapping objects // API name (keep as is) api_name="/predict" ]); ```🧑💻 Napari Plugin
Find our Napari Plugin here: https://github.com/FZJ-INM1-BDA/celldetection-napari
Find out more about Napari here: https://napari.org
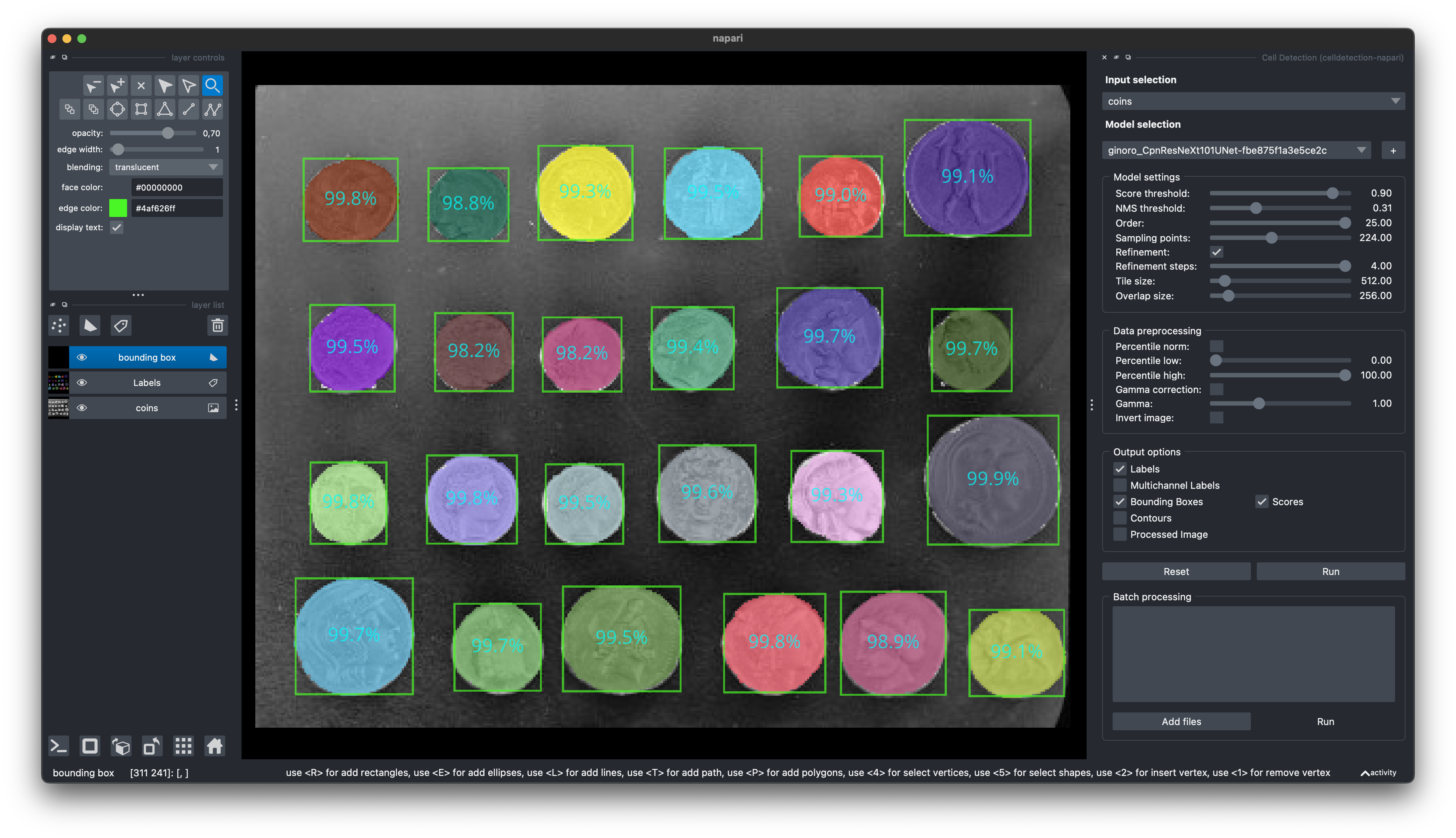 You can install it via pip:
You can install it via pip:
pip install git+https://github.com/FZJ-INM1-BDA/celldetection-napari.git🏆 Awards
- NeurIPS 2022 Cell Segmentation Challenge: Winner Finalist Award
📝 Citing
If you find this work useful, please consider giving a star ⭐️ and citation:
@article{UPSCHULTE2022102371,
title = {Contour proposal networks for biomedical instance segmentation},
journal = {Medical Image Analysis},
volume = {77},
pages = {102371},
year = {2022},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2022.102371},
url = {https://www.sciencedirect.com/science/article/pii/S136184152200024X},
author = {Eric Upschulte and Stefan Harmeling and Katrin Amunts and Timo Dickscheid},
keywords = {Cell detection, Cell segmentation, Object detection, CPN},
}
