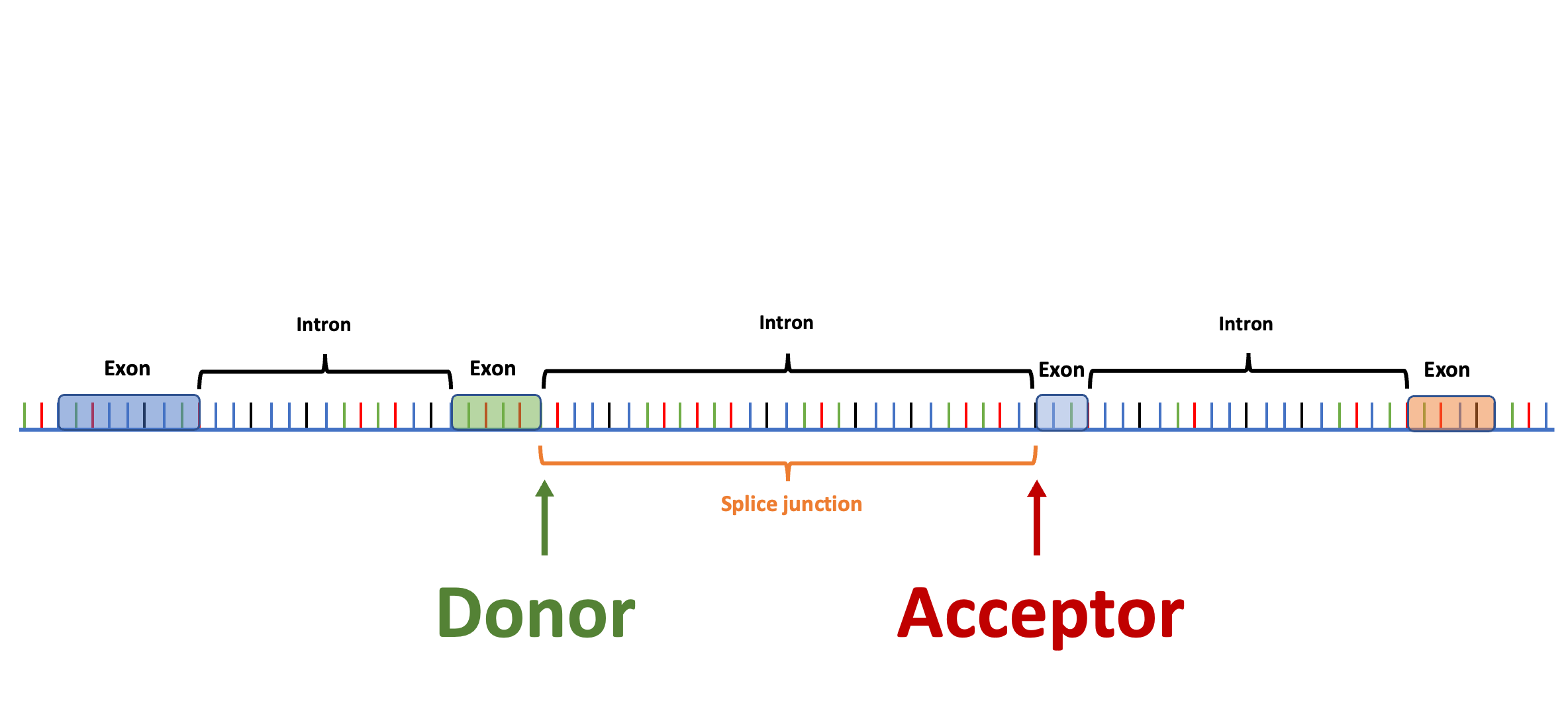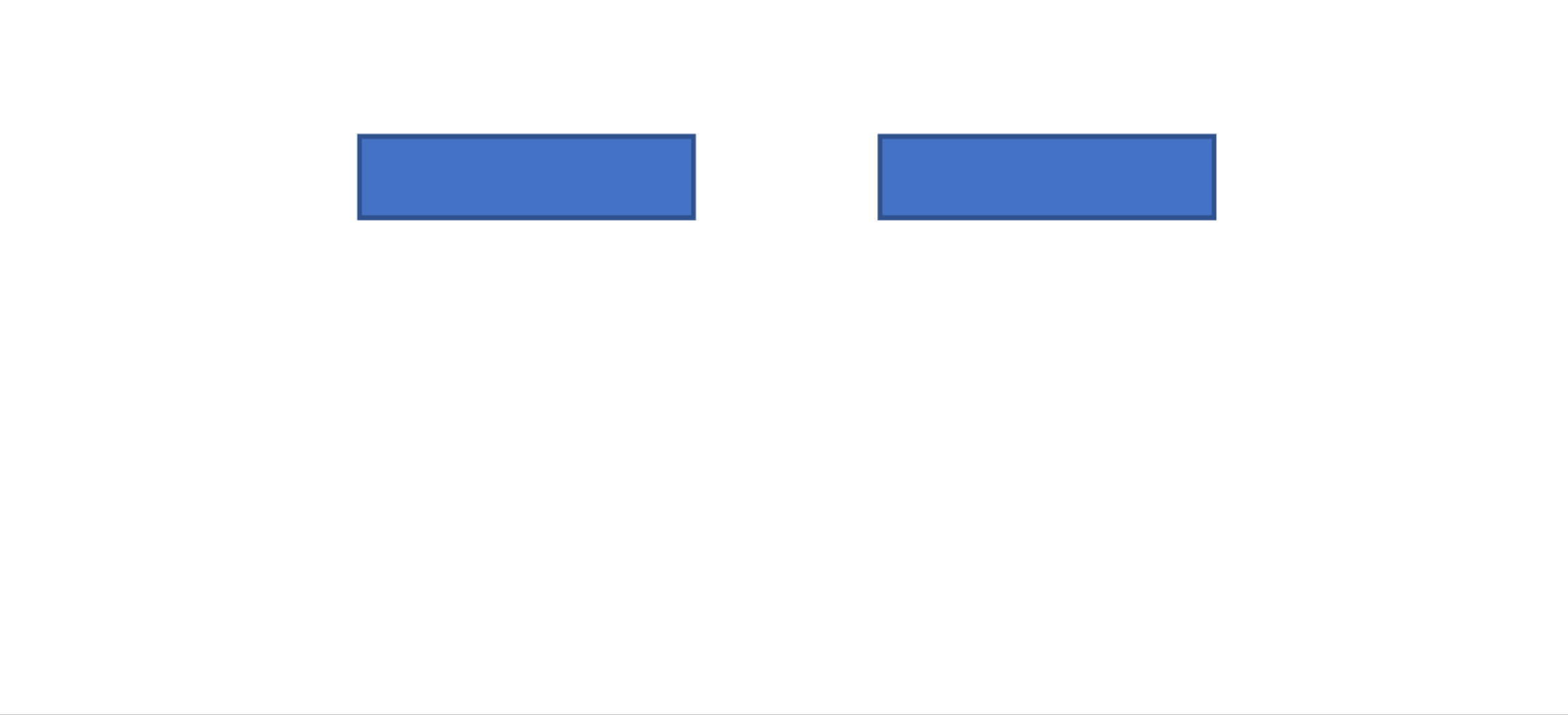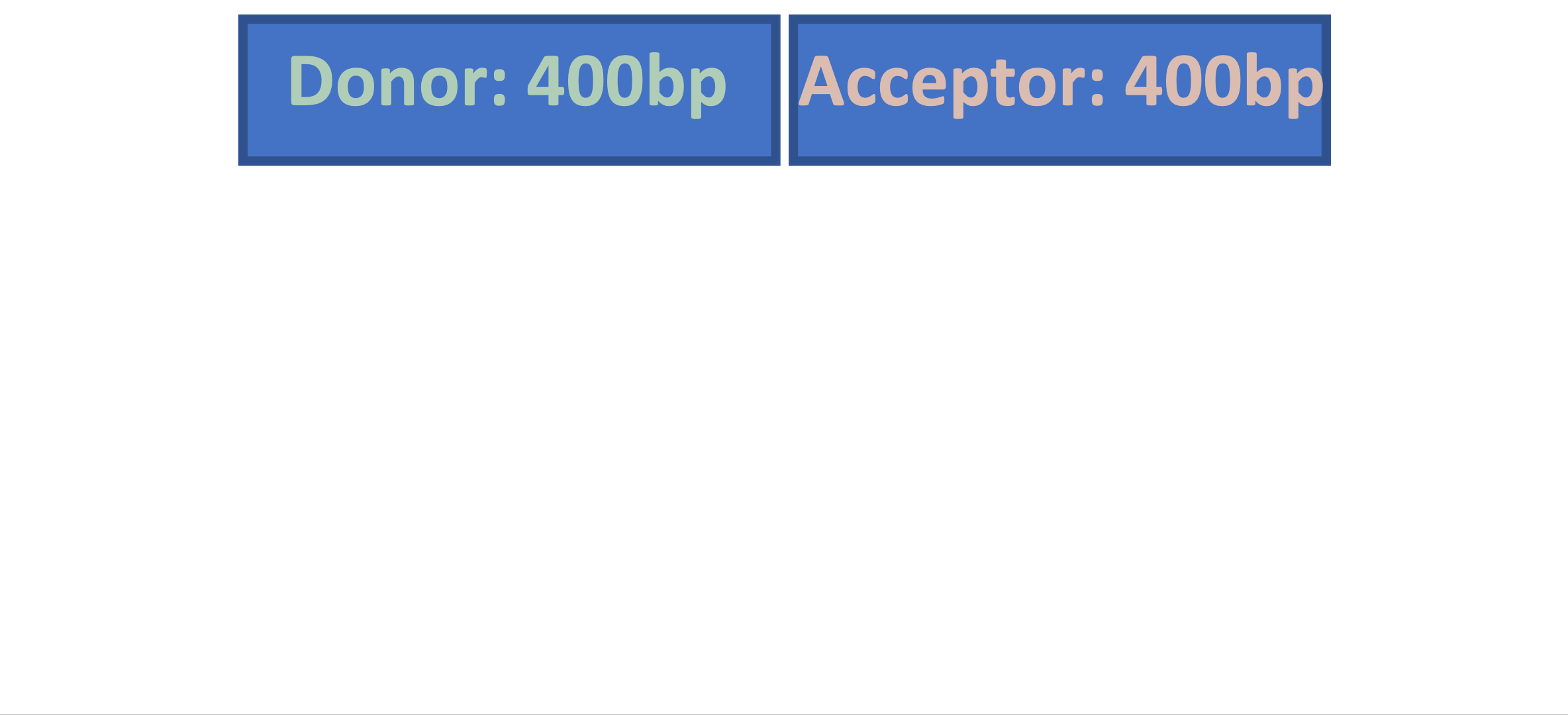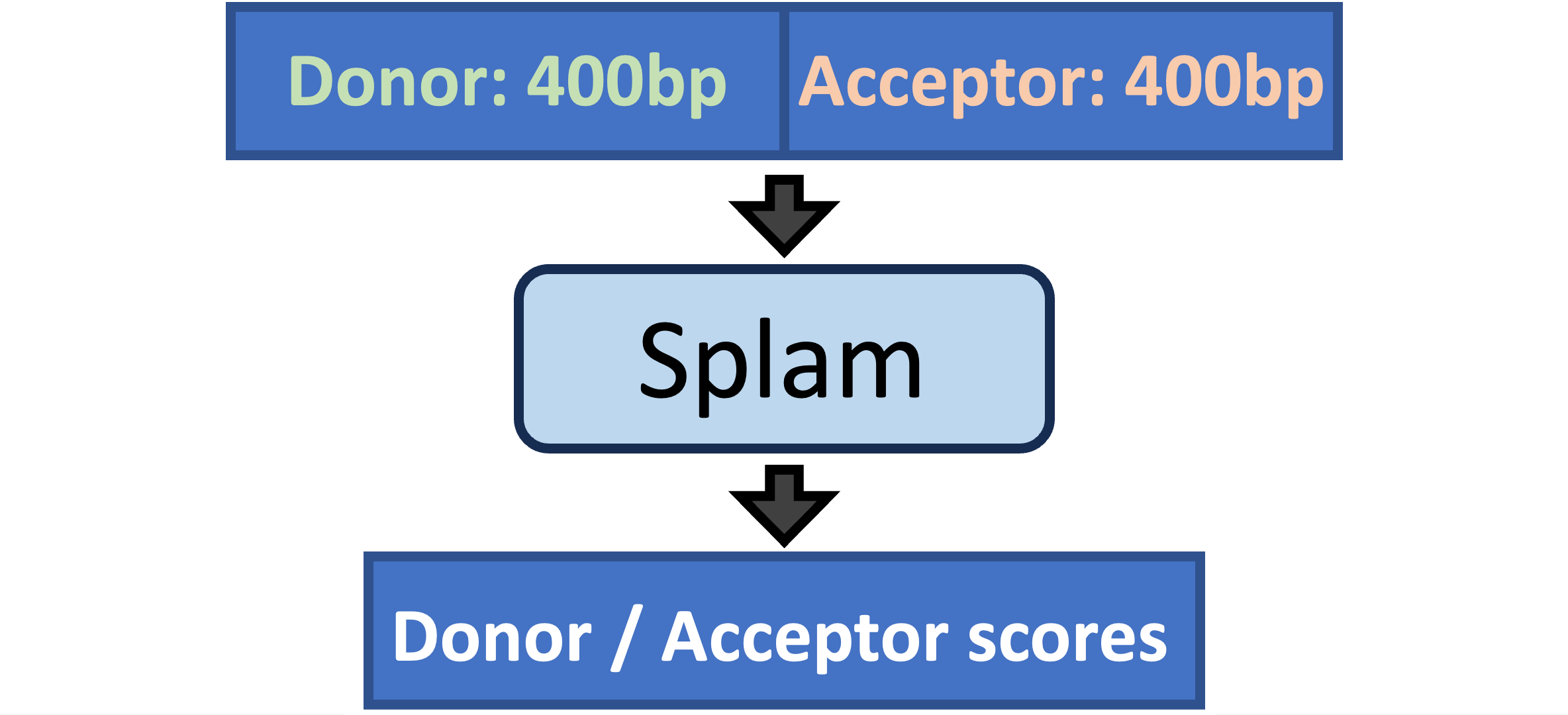
Splam is a splice site predictor utilizing a deep residual convolutional neural network for fast and accurate evaluation of splice junctions solely based on 400nt DNA sequences around donor and acceptor sites.
Why Splam❓#
- We need a tool to evaluate splice junctions & spliced alignments. Thousands of RNA-Seq datasets are generated every day, but there are no tools available for cleaning up spurious spliced alignments in these data. Splam addresses this problem!
- Splam-cleaned alignments lead to improved transcript assembly, which, in turn, may enhance all downstream RNA-Seq analyses, including transcript quantification, differential gene expression analysis, and more.
Who is it for❓#
If you are (1) doing RNA-Seq data analysis or (2) seeking a trustworthy way to evaluate splice junctions (introns), then Splam is the tool that you are looking for!
What does Splam do❓#
There are two main use case scenarios:
-
Improving your alignment file. Splam evaluates the quality of spliced alignments and removes those containing spurious splice junctions. This significantly enhances the quality of downstream transcriptome assemblies [Link].
-
Evaluating the quality of introns in your annotation file or assembled transcripts [Link].
Documentation#
📒 The full user manual is available here
Table of contents#
## Installation# Splam is on [PyPi](https://pypi.org/). This is the easiest installation approach. Check out all the releases [here](https://pypi.org/manage/project/splam/releases/). ```bash $ pip install splam ``` You can also install Splam from source ```bash $ git clone https://github.com/Kuanhao-Chao/splam --recursive $ cd splam/src/ $ python setup.py install ```
## Quick Start# Running Splam is simple. It only requires three lines of code! See these examples on Google Colab: [](https://colab.research.google.com/github/Kuanhao-Chao/splam/blob/main/notebook/splam_example.ipynb) ### Example 1: clean up alignment files (`BAM`) ``` bash $ cd test # Step 1: extract splice junctions in the alignment file $ splam extract -P SRR1352129_chr9_sub.bam -o tmp_out_alignment # Step 2: score all the extracted splice junctions $ splam score -G chr9_subset.fa -m ../model/splam_script.pt -o tmp_out_alignment tmp_out_alignment/junction.bed #Step 3: output a cleaned and sorted alignment file $ splam clean -o tmp_out_alignment ``` ### Example 2: evaluate annotation files / assembled transcripts (`GFF`) ``` bash $ cd test # Step 1: extract introns in the annotation $ splam extract refseq_40_GRCh38.p14_chr_fixed.gff -o tmp_out_annotation # Step 2: score introns in the annotation $ splam score -G chr9_subset.fa -m ../model/splam_script.pt -o tmp_out_annotation tmp_out_annotation/junction.bed #Step 3: output statistics of each transcript $ splam clean -o tmp_out_annotation ``` ### Example 3: evaluate mouse annotation files (`GFF`) ``` bash $ cd test # Step 1: extract introns in the annotation $ splam extract mouse_chr19.gff -o tmp_out_generalization # Step 2: score introns in the annotation $ splam score -G mouse_chr19.fa -m ../model/splam_script.pt -o tmp_out_generalization tmp_out_generalization/junction.bed # Step 3: output statistics of each transcript $ splam clean -o tmp_out_generalization ```
## Scripts for Splam model training & analysis# All the scripts for Splam training and data analysis are in [this GitHub repository](https://github.com/Kuanhao-Chao/splam-analysis-results).
## Citation# Kuan-Hao Chao*, Alan Mao, Steven L Salzberg, Mihaela Pertea*, "Splam: a deep-learning-based splice site predictor that improves spliced alignments ", bioRxiv 2023.07.27.550754, doi: [https://doi.org/10.1101/2023.07.27.550754](https://doi.org/10.1101/2023.07.27.550754), 2023





