PIA - Protein Inference Algorithms
PIA is a toolbox for MS based protein inference and identification analysis.
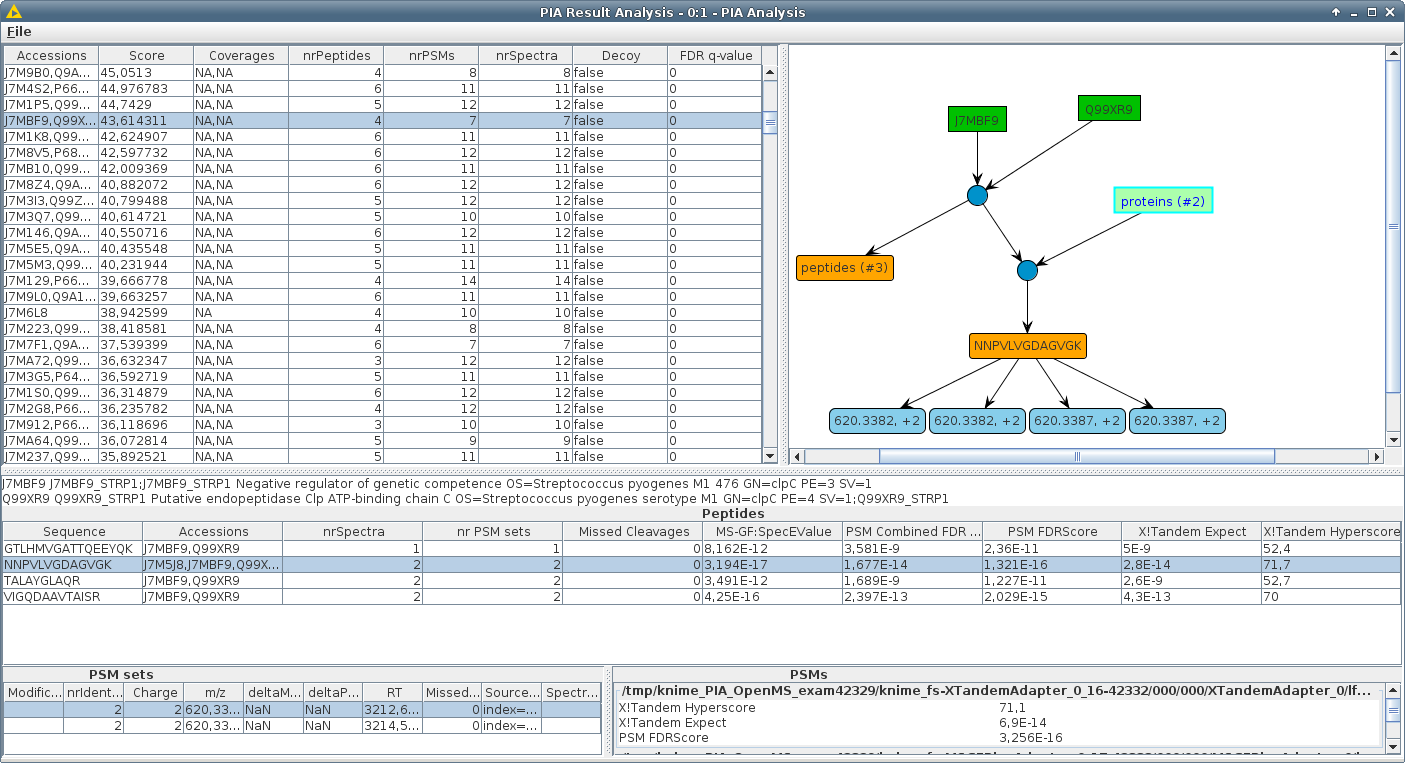
PIA allows you to inspect the results of common proteomics spectrum identification search engines, combine them seamlessly and conduct statistical analyses. The main focus of PIA lays on the integrated inference algorithms, i.e. concluding the proteins from a set of identified spectra. But it also allows you to inspect your peptide spectrum matches, calculate FDR values across different search engine results and visualize the correspondence between PSMs, peptides and proteins.

PIA in a nutshell
Most search engines for protein identification in MS/MS experiments return protein lists, although the actual search yields a set of peptide spectrum matches (PSMs). The step from PSMs to proteins is called "protein inference". If a set of identified PSMs supports the detection of more than one protein in the searched database ("protein ambiguity"), usually only one representative accession is reported. These representatives may differ according to the used search engine and settings. Thus the protein lists of different search engines generally cannot be compared with one another. PSMs of complementary search engines are often combined to enhance the number of reported proteins or to verify the evidence of a peptide, which is improved by detection with distinct algorithms.
We developed an algorithm suite written in Java, including fully parametrisable KNIME nodes, which combine PSMs from different experiments and/or search engines, and reports consistent and thus comparable results. None of the parameters, like filtering or scoring, are fixed as in prior approaches, but held as flexible as possible, to allow for any adjustments needed by the user.
PIA can be called via the command line (also in Docker containers) or in the workflow environment KNIME, which allows a seamless integration into OpenMS workflows.
Download
For the command line you can download the latest released version
using Conda (respectively Bioconda)

PIA is also integrated into KNIME. You can easily install it from the trusted community contributions repository, which is available in all newer KNIME versions. Please use always the newest version of KNIME before submitting any bugs or issues. More information on how to install and run PIA inside KNIME can be found in the wiki about PIA in KNIME.
Tutorial
The tutorial as PDF can be downloaded here, the required data are available here and the workflows here (all data is also available in the tutorial repository at https://github.com/mpc-bioinformatics/pia-tutorial/).
Documentation
For further documentation please refer to the Wiki (https://github.com/medbioinf/pia/wiki) on github.
Problems, Bugs and Issues
If you have any problems with PIA or find bugs and other issues, please use the issue tracker of GitHub (https://github.com/medbioinf/pia/issues).
Citation and Publication
If you found PIA useful for your work, please cite the following publications:
https://www.ncbi.nlm.nih.gov/pubmed/25938255
https://www.ncbi.nlm.nih.gov/pubmed/30474983
Funding
The development of PIA is funded by ELIXIR / de.NBI, the German Network for Bioinformatics Infrastructure.
Authors of PIA
The programming work on PIA was performed by Julian Uszkoreit (Ruhr University Bochum, Medical Bioinformatics), and Yasset Perez-Riverol (European Bioinformatics Institute (EMBL-EBI), Cambridge)
